The Yu Lab at BCH

The Yu Lab has demonstrated that MAPK8IP3 RNA processing is naturally inefficient, creating opportunities to turn the gene on or off in different cell types, or boost gene expression with ASOs. The Yu Lab has identified several RNA processing inefficiencies in the MAPK8IP3 locus in fibroblasts, astrocytes and iPSC-derived neurons and developed antisense oligonucleotides that increase correctly spliced MAPK8IP3 transcripts in all cell types. The Yu Lab also demonstrated significantly increased protein levels in fibroblasts and astrocytes.
About Dr Yu
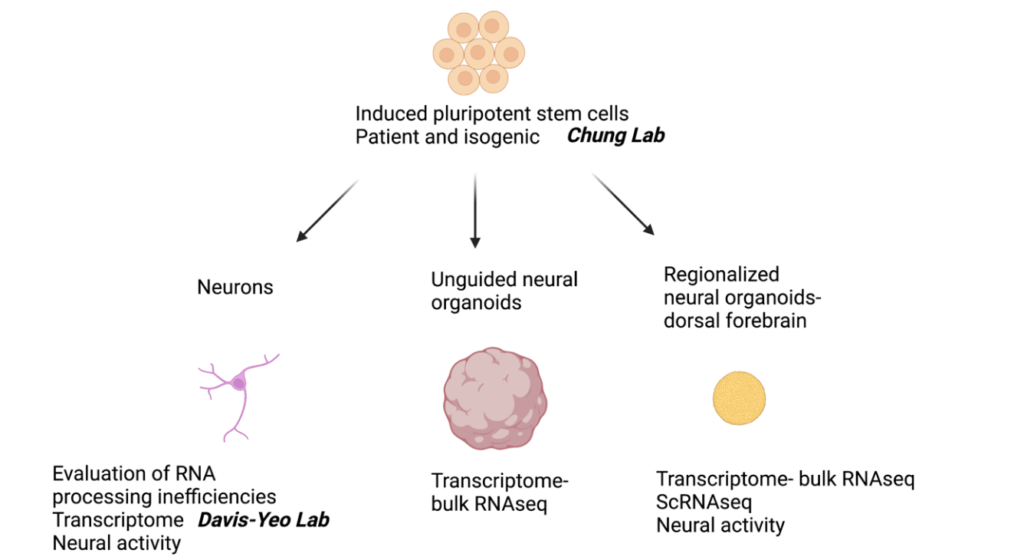
Figure 1. Summary of experimental models used in the Yu Lab to study MAPK8IP3 mutations
The Yu Lab has successfully generated brain organoids from MAPK8IP3 R578C/+ iPSCs (and isogenic controls) and MAPK8IP3 M543del/+ iPSCs, allowing us to model brain development in multiple cell lineages (Figure 1 and Figure 2). Organoids morphologically developed as expected and they expressed neural and neuroprogenitor markers tested by immunohistochemistry and gene expression analyses. Further QC steps included generation of single cell RNAseq data to confirm expression of relevant marker genes and demonstrate reproducible cell type proportions. MAPK8IP3 organoids exhibit abnormal gene signatures that provide leads for development as candidate biomarkers.
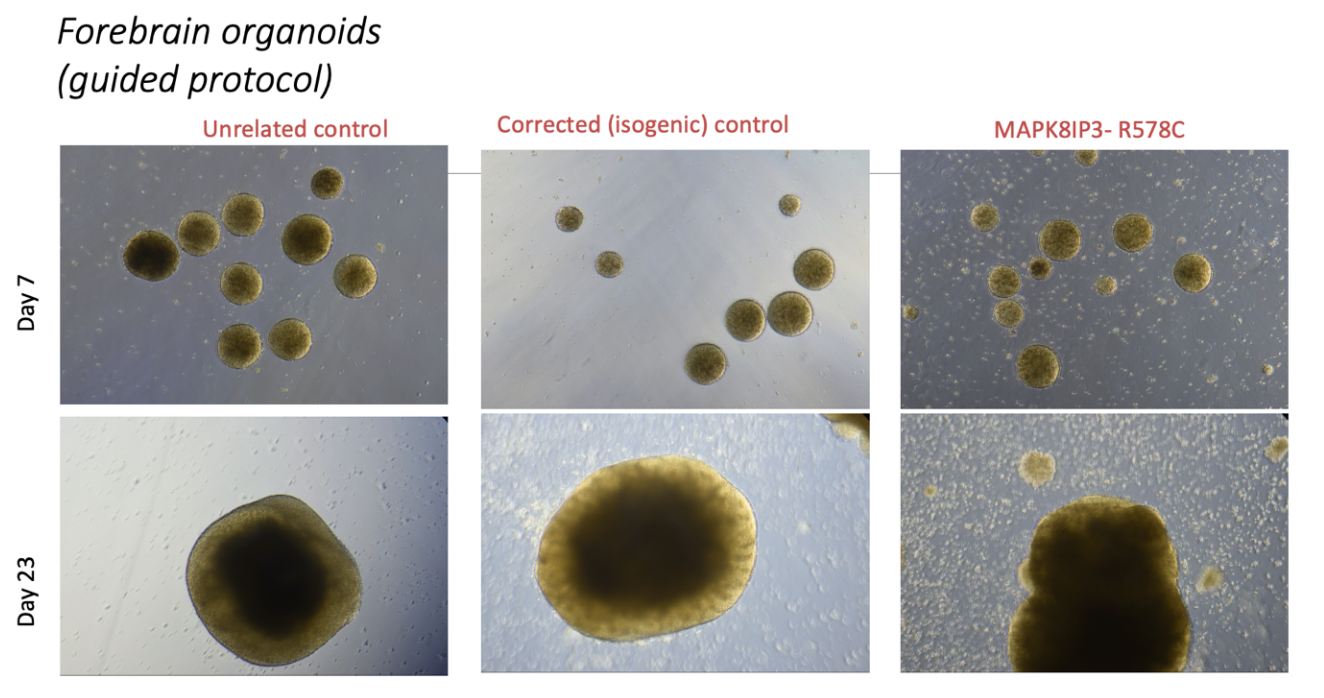
Figure 2. MAPK8IP3 and control organoids are shown at different days of differentitation.
